At the Howard Hughes Medical Institute, we believe in the power of individuals to advance science through research and science education, making discoveries that benefit humanity.
The University of Oregon is a public flagship research university in Eugene, Oregon. Founded in 1876, the institution's 295-acre campus is located in the southern Willamette Valley.
Research
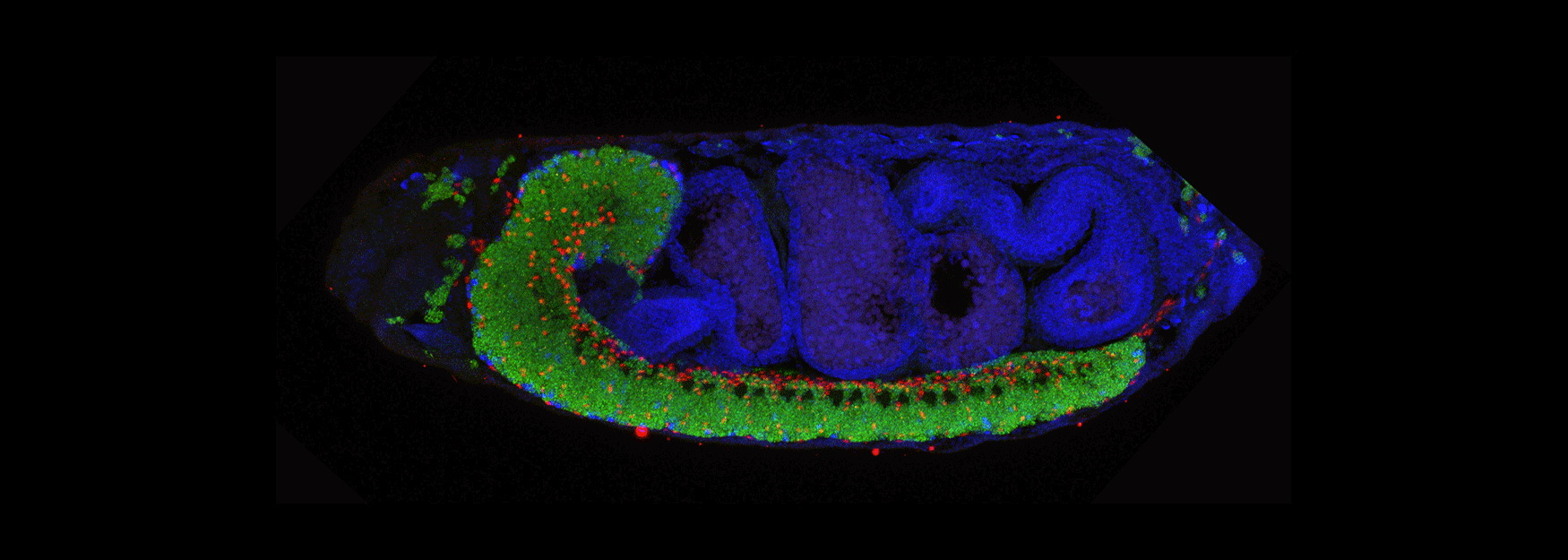
We are interested in how Drosophila neural progenitors (neuroblasts) generate neuronal diversity, and how diverse neurons establish precise neural circuits to generate motor behavior. All motor behaviors, from simple reflexes to highly choreographed movements, depend on the establishment and function of neural circuits in the brain and spinal cord. Disruption of motor circuits in disease or injury can have devastating effects, and thus a major goal of neuroscience is to devise treatments to restore circuit function. Understanding of how neural circuits originally assemble could provide a roadmap for re-building neural circuits.
Our lab is interested in four related research areas:
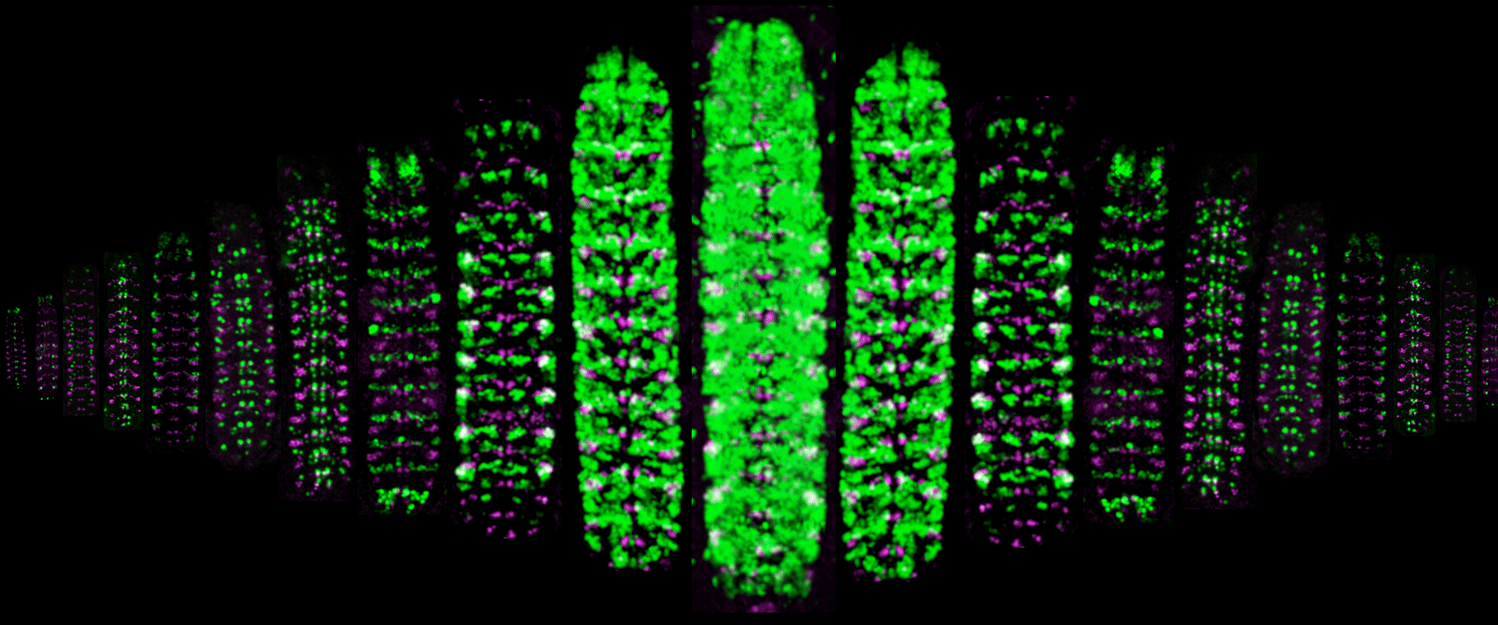
(1) How are spatial and temporal cues integrated to specify neuronal identity? Neuroblast identity is determined by transient spatial cues; neuroblast progeny are determined by a temporal transcription factor (TTF) cascade. How are these molecular inputs integrated? How does transient exposure to spatial and temporal cues lead to heritable neuronal identity: via a gene expression cascade or by modifying chromatin?
(2) How do TTFs generate neuronal diversity? What is the "timer" that produces sequential TTF expression in embryonic or larval neuroblasts? Does the timer require intrinsic or extrinsic input? How does TTF expression in neuroblasts lead to heritable neuronal identity? Larval brain "type II" neuroblasts generate the adult central complex (used for celestial navigation); both the neuroblasts and their INP progeny undergo temporal patterning. How are neuroblast and INP temporal cascades are integrated?
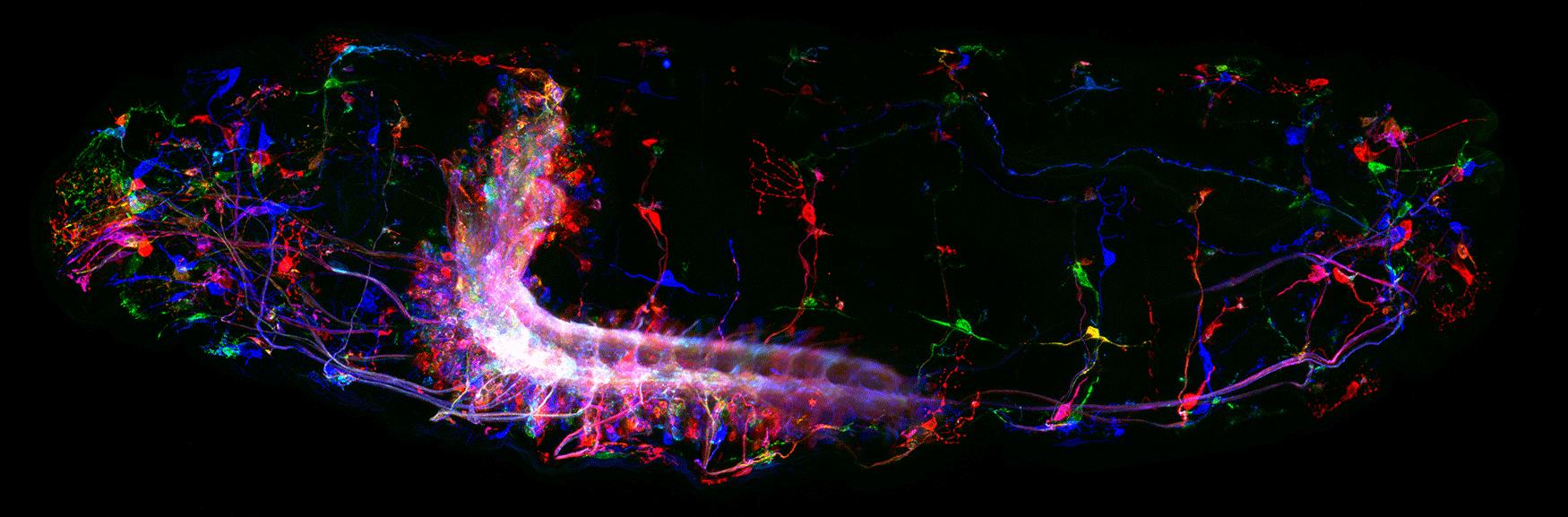
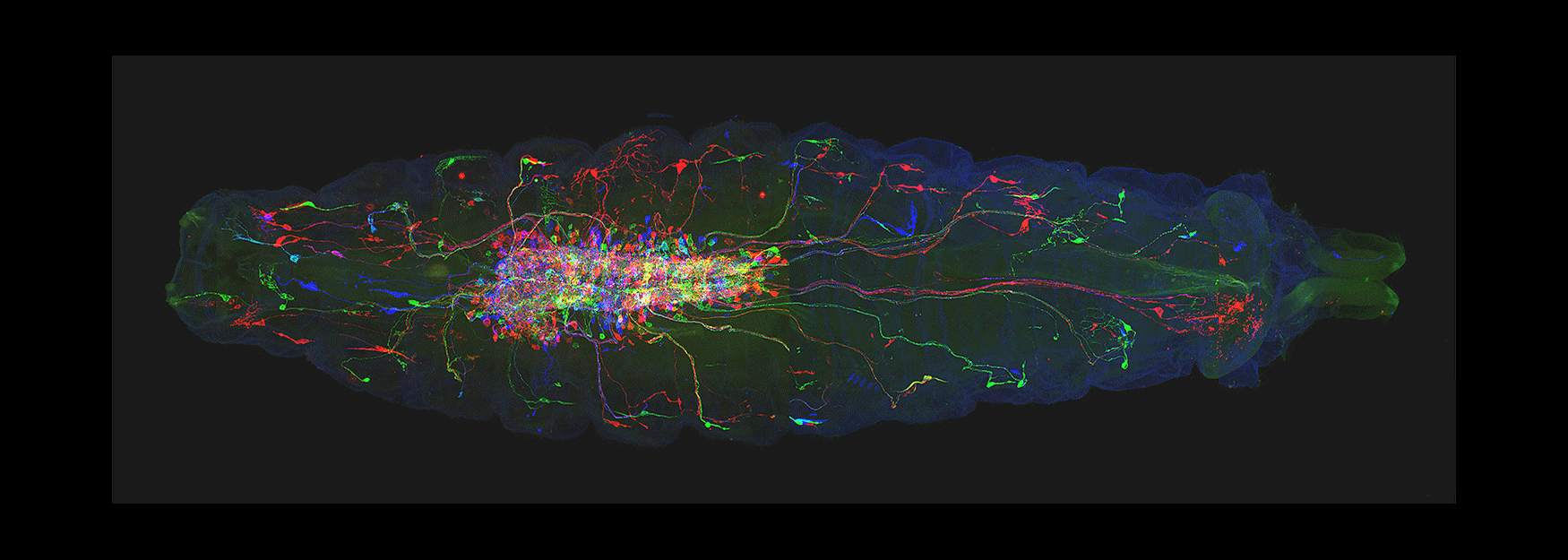
(3) How are developmental mechanisms used to establish neural circuits? We use two model systems: larval motor circuits and adult navigation circuits. Are clonally-related neurons from a single neuroblast preferentially connected? Are early-born neurons from multiple neuroblasts preferentially connected? Are the same developmental mechanisms used to establish neural circuits also used to maintain neural circuits?
(4) How are circuits maintained from larval to adult? We have shown that the "moonwalker descending neuron" (MDN) is present from embryo to adult, and can induce backward locomotion in both larvae and adults. How does MDN drive backward locomotion in a legless larva and a limbed adult? What does the MDN circuit look like in larvae and adults? Is there a persistent core circuit that is maintained? Are the mechanisms that establish MDN circuits in the embryo the same as those establishing MDN circuits in the adult?